Released: 7-Nov-2014 1:00 PM EST
Source Newsroom: Rockefeller University
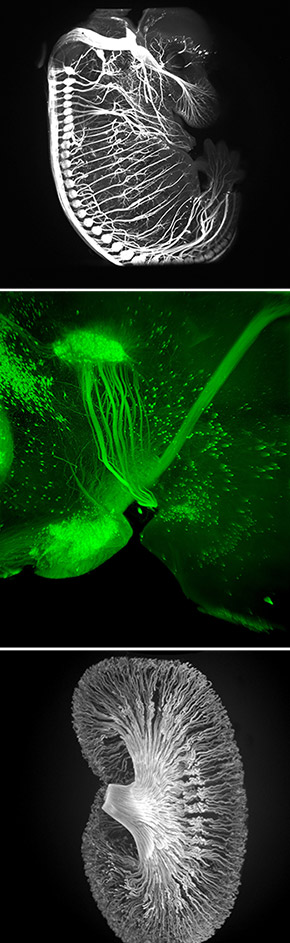
Newswise — In a significant technical advance, a team of neuroscientists at The Rockefeller University has devised a fast, inexpensive imaging method for probing the molecular intricacies of large biological samples in three dimensions, an achievement that could have far reaching implications in a wide array of basic biological investigations.
The new method, called iDISCO, optimizes techniques for deep tissue immunolabeling and combines them with recent technological innovations in tissue clearing and light sheet microscopy to achieve unprecedented deep labeling and imaging of molecular structures in the brain, the kidney, and other organs and tissues in experimental settings. A detailed report on iDISCO is in the November 6 issue of the journal Cell.
“What we did was optimize many different parameters of several existing techniques to create this powerful new labeling method,” says Marc Tessier-Lavigne, Rockefeller president, Carson Family Professor, head of the Laboratory of Brain Development and Repair, and senior author on the new study. “These optimization efforts paid off in spades, dramatically extending our ability to visualize molecular structures deep in intact complex tissues like the brain. Although developed in our laboratory to help us pursue neuroscience questions, we believe this new method will provide biological researchers in many disciplines with an important new tool for advancing their work.”
When executed as part of a coordinated protocol, the advance creates a powerful method for imaging molecular structures deep in tissues that is much simpler and more rapid than previous approaches used by biological researchers.
“For us as neuroscientists, one of the big applications of this new imaging method has been the ability to visualize axonal pathways in the developing and adult brain,” says Nicolas Renier, a postdoctoral associate in the Tessier-Lavigne laboratory and co-first author on the study. “We were surprised at just how well we were able to image these detailed structures in the context of the whole brain.”
“The fact that we can now visualize neural circuit formation in larger embryos allows us to study the developing nervous system when it is more well formed,” says Zhuhao Wu, also a postdoctoral associate in the Tessier-Lavigne laboratory and co-first author on the study. “This opens entire new avenues to our research.”
The team focused on three techniques, as they sought to maximize the effectiveness of deep tissue immunolabeling and combine it with innovations by other scientists in tissue clearing and light sheet microscopy.
Clearing is a chemical treatment process that renders biological samples transparent, enabling light to reach deep inside them, and it is in this area particularly that new frontiers have been established in a number of laboratories recently, making it possible to peer deeper into samples than ever before.
Immunolabeling is a longstanding laboratory technique for tagging molecules of interest in biological samples with selected antibodies, but usually in relatively small or thin samples. In this area, the Rockefeller group demonstrated their ability to introduce 28 widely used research antibodies much more deeply and more rapidly into a diversity of samples than had previously been possible. Immunolabeling differs from another commonly used technique in which fluorescent reporter molecules are introduced into a tissue transgenically. Immunolabeling instead attaches tags to endogenous molecules in a study sample.
“Being able to visualize molecules introduced transgenically by the experimenters is a very valuable tool,” says Tessier-Lavigne. “What drove us, however, was a desire to complement that tool with a greater ability to look at endogenous molecular processes. In this area, our new labeling method breaks a barrier we weren’t necessarily expecting to be able to break when we set out on this project.”
Light sheet microscopy has the power to scan whole organs or large tissues in relatively short amounts of time at very high resolutions. The data set captured is so detailed that it can be translated either into images offering a comprehensive view of the entire sample or a closer look at smaller structures within the sample.
Renier and Wu are co-lead authors on the Cell study, and Tessier-Lavigne is senior author. Their coauthors are: David I. Simon, Jing Yang and Pablo Ariel, also in the Tessier-Lavigne laboratory.
